From the Depths of Literature: How Large Language Models Excavate Crucial Information to Scale Drug Discovery
Excavating crucial information about potential drug targets and molecules from the depths of the scientific literature is a time-consuming task for drug discovery teams. Large Language Models offer fast, scalable, and accurate solutions for biotechs. Here is how, in three concrete examples.
Biotech and Pharma are information-heavy industries where better access to knowledge about diseases, drugs, and their targets is crucial for decision-making. But the ever-expanding volume of published research is both a blessing and a curse. Valuable knowledge often remains buried deep within the scientific literature, scattered across publications, conference abstracts, patents, or press releases. Sometimes, the key pieces of information needed to make a critical decision on a target can be hidden in a number of convoluted sentences from publications dating back to 1995 to 1999.
While humans are adept at navigating this complex information landscape at a small scale, their time is limited and costly. More often than not, the resources available to R&D teams are no match for the amount of potentially relevant information. Somewhat paradoxically, as the body of scientific knowledge is growing, the share of information drug discovery teams can effectively use may actually be decreasing.
Therefore, computer scientists, machine learning and NLP experts have struggled for decades to develop models that extract and summarise relevant information from scientific literature in a way that could truly aid scientists in making well-grounded decisions. Up to now, despite significant investment, progress has been limited.
With the recent breakthrough of Large Language Models (LLMs) underlying products like ChatGPT, the game has changed. These powerful language models can process text in a manner that closely resembles human understanding, in seconds not hours.
Here we present three valuable applications of LLMs that can directly benefit biotech and pharma companies in bridging the gaps in scientific knowledge faster to make more efficient choices in drug development:
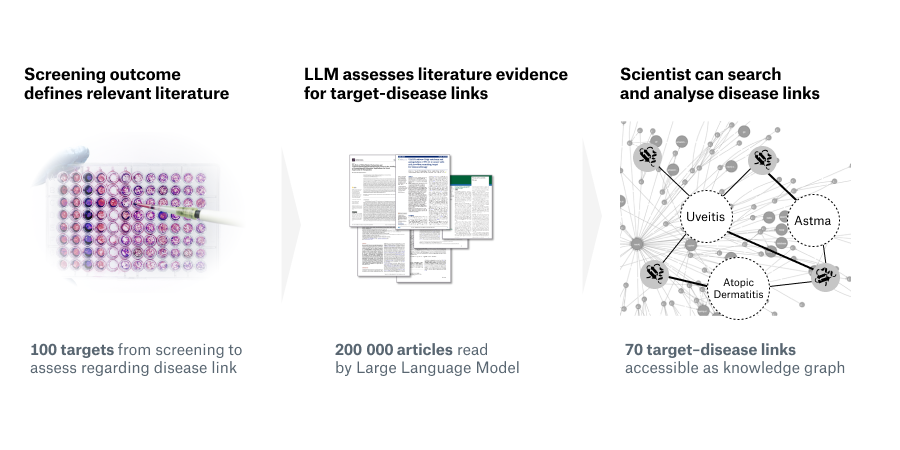
Prioritising disease-relevant targets from screens
Assessing target-related adverse event profiles
Identifying biomarkers for a new target
Finding target-disease links
As the number of new drugs continues to grow, the range of addressable targets has historically been limited by the structural dependencies of classical small molecule drugs. However, the landscape is rapidly evolving with recent advancements in pharma and biotech, opening up a vast space of potential drug targets. First of all, novel therapeutic modalities like PROTACs, innovative biologics, and nucleotide-based therapeutics offer new ways to address targets; and secondly, new scientific insights into the so-called “dark” regions of the genome and proteome, expand the target space itself.
But amidst this abundance of potential targets, the key question is identifying which ones truly influence relevant disease contexts, such as acting as apical regulators in disease-specific pathways. Many targets play roles in multiple different diseases, such as members of the interleukin family that act in signalling pathways of immune-related and infectious diseases, as well as cancer.
For example, if we wanted to discover novel modulators of the IL-6 pathway and their links to various immune-related diseases? Conducting a quick PubMed search using the keywords "IL-6 inhibitor" and "disease" yields over 14,000 research papers published between 1986 and 2023, all of which may contain valuable information related to diseases.
Another way of identifying disease-relevant targets could be to start with the results of a high-throughput target screen. This screen would identify around 100 additional targets whose knockdown demonstrated similar effects as the IL-6 inhibitor in regulating cytokine levels. Again, a quick PubMed search yields more than 200,000 papers that scientists would need to consider when prioritising those targets.
It is here where LLM’s ground-breaking capabilities are changing the game of drug discovery. In contrast to previous NLP or search engine technology, LLMs can accurately retrieve crucial information from vast amounts of data, such as:
The link between a target and a disease
The role that the target plays in the disease, e.g. apical regulator or downstream effector
The confidence in the link between the target and the disease
Once extracted from the literature, this information can be easily explored in scoring tables or in knowledge graphs, which highlight the links between targets and diseases.
Assessing Adverse events
When it comes to decision-making regarding targets and molecule classes, whether for internal portfolio prioritisation or when conducting due diligence on prospective partners, understanding the potential for adverse events related to targets, compound classes, and modalities is essential. But the reporting of such data in the scientific literature can be convoluted and challenging to interpret, even for humans, due to differing nomenclatures, cutoff values, dosing schemes, drug combinations, indications, and patient populations. Moreover, crucial information is often buried in conference abstracts or supplementary tables.
Take, for instance, drugs acting on the receptor tyrosine kinase EGFR, for which the FDA AERS database alone contains 27,123 reported adverse events between 2004 and 2018. To make sense of the adverse event profiles of EGFR drugs reported in scientific literature, researchers are faced with more than 2,500 articles on PubMed, requiring a thorough analysis to extract and understand the set of reported adverse events.
Rather than researchers struggling to piece together scattered information, LLMs can provide a more efficient approach. By feeding relevant literature into the LLM, it can generate a comprehensive table with unified nomenclature, organising all reported adverse events based on the compound, affected organ or tissue, and severity. Moreover, the model can also incorporate data on on-target versus off-target toxicity and identify specific red flag toxicities.
This allows scientists to make a quick and fair prediction of a candidate’s inherent tox profiles, making sure that biotech decision-makers can compare apples and apples. This approach not only saves scientists valuable time but can also help to uncover vital patterns of potential drug- or compound-related risks that might have been overlooked by a human reader.
Discovering Biomarkers
As treatments for various diseases shift towards personalised approaches, biomarkers defining patient populations and their response to specific drugs have become more crucial than ever. Understanding the disease pathway in which a target operates can lead to the discovery of novel downstream and upstream regulators that could serve as biomarkers in preclinical experiments. However, when evaluating multiple targets, each potentially acting in diverse pathways, the complexity of the information to be assessed can quickly escalate. Furthermore, biomarkers might not only emerge from the most obvious pathways but also from parallel signalling routes that may initially seem disconnected. This complexity further intensifies when assessing drug combinations.
For instance, when searching for drug candidates that can enhance the response to immune checkpoint inhibitors, there are already over 5,000 papers discussing potential biomarkers, not to mention numerous articles listing potential biomarkers that are not yet on the scientists' radar.
In this context, LLMs can help scientists navigate the complexity of potential biomarkers and their targets. They can not only extract relationships between targets and their upstream and downstream regulators but also extract the directionality, context and potential size of the effect. Moreover, LLMs could also highlight where the biomarker might be discovered, e.g. tumour vs. blood and identify suitable assays to measure the biomarker. This makes LLMs invaluable tools for identifying biomarkers for novel drugs and drug combinations.
Conclusion
After decades of glorified keyword search tools, large language models truly change the nature of how we can deal with scientific literature. LLM-based tools help scientists to quickly extract and aggregate vital information from complex scientific literature and inform decision-making in biotech and pharma.
At idalab, we build LLM-based tools, which are tailored to our clients’ unique research questions. By refining and optimising input data, we anchor the model with relevant information and customise output formats to fit our clients' specific research areas, target classes, and development stages. Building on our established platform, data infrastructure and expertise, the whole process only takes a couple of weeks.
Book a discovery call
In our free 30-minute 1:1 discovery calls, a member of our team will be happy to answer your questions, share lessons learned and discuss how to address your specific needs.
Further reading
Raising the efficiency floor and innovation ceiling with generative AI in drug discovery (Brian Buntz in Drug Discovery and development) introduces recent innovative uses of generative AI in the biotech industry
Large language models encode clinical knowledge (Karan Singhal et. al in Nature) introduces ways in which Large Language Models can be used to parse medical data
Drug discovery companies are customizing ChatGPT: here’s how (Neil Savage in Nature) discusses the various ways in which biotechs are putting Large Language Models to use